Hypoxia-induced molecular alterations and druggable targets in triple negative breast cancer cell lines - HipTNBC
Aim
The aim of the project is to identify and to verify molecular alterations and druggable targets associated with acute, cycling, and chronic hypoxia in TNBC.
Description
Graphical abstract
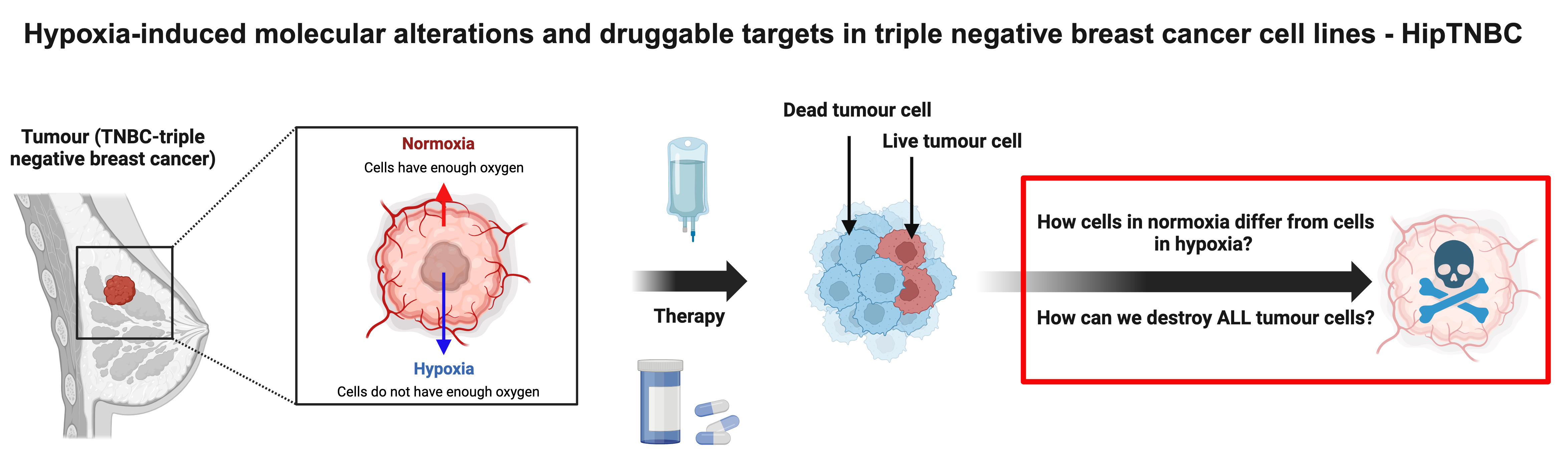
Methods
Hypoxia-induced molecular alterations (somatic genomic alterations; mRNA, miRNA, protein expression and DNA methylation) and druggable targets (e.g., BH3 mimetics) based on mitochondrial apoptotic dependencies in TNBC will be identified by bioinformatics analysis of publicly available multi-omics data sets, using hypoxia-related mRNA signatures. Molecular alterations and druggable targets identified during this and baseline and dynamic BH3 profiling analysis will be validated in TNBC reference cell lines exposed to acute hypoxia, cycling hypoxia and chronic hypoxia.
Outcomes
The outcomes of this research will be potential tumour-hypoxia related indicators and list of potential therapeutic agents for development of personalized therapy field. In a long-term perspective, the project will contribute to improvements in the management, survival, and health-related quality of life of TNBC patients.
The results of the project will be presented in original scientific articles in journals listed in Web of Science or SCOPUS, as well as in international scientific conferences. The results will be used for future research projects as well as for the PhD thesis defence of the PhD student(s) involved in the project.
Scientific objectives
- To perform bioinformatics analysis in publicly available TNBC multi-omics data sets (TCGA, METABRIC) for identification of novel acute continuous and cycling hypoxia-associated molecular alterations (potential biomarkers) and relevant druggable targets.
- To verify molecular alterations identified in bioinformatics analysis, as well as 15-gene expression continuous acute hypoxia signature and cycling hypoxia signature in the panel of triple negative breast cancer-reference cell lines (TNBC-RCLs) exposed to various modes of hypoxia.
- To explore the transcriptomic and proteomic expression status of various isoforms of HIFs (HIF-1, HIF2, HIF-3) during exposure of TNBC-RCLs to various modes of hypoxia.
- To identify mitochondrial apoptotic dependencies induced by various hypoxia conditions for identification of druggable targets for BH3 mimetics.
- To explore the effects of various modes of hypoxia on the sensitivity of TNBC-RCLs to the pharmacological agents identified in the bioinformatics and mitochondrial apoptotic priming dependency analysis.
Benefit for society: the results of the project will help to improve future treatment choices, care, survival, and health-related quality of life for patients with aggressive and difficult-to-treat breast cancer.
Issues the project will address: 'hypoxia' refers to conditions in which oxygen does not reach the cells in sufficient quantities. Normal cells usually die, but malignant cells can adapt to these conditions and survive. The project will find out how cells that survive hypoxia differ from other cells and how to destroy them effectively.
Possible solutions: To find out how hypoxic survivors (cells) differ from dead cells; we will grow breast cancer cells under low oxygen conditions and identify which gene programmes are triggered by these conditions and how cancer cells escape death.
Olesja Rogoza presents the research project at the young scientist session of the PMNET Forum 2024
Project implementors
RSU cancer cell research group:
- Dr.biol. Inese Čakstiņa-Dzērve – project leader
- Dipl.pharm. Valdis Pirsko - technologist
- Dipl.pharm. Ingrīda Mitre - researcher
- Msc.biol. Olesja Rogoza – PhD student




